Introduction to toolStability
Tien-Cheng Wang
2025-05-14
Source:vignettes/toolStability.Rmd
toolStability.RmdOverview
The package toolStability is part of the publication
from (Wang, Casadebaig,
& Chen, 2023). The package is a collection of functions
which implements eleven methods for describing the
stability of a trait in terms of
genotype and environment.
The goal of this vignette is to introduce users to these functions
and get started in analyzing dataset with interaction between
genotype and environment.
Further analysis for original publication using this package can be found in the link ( click).
Installation
The package can be installed using the following functions:
# Install from CRAN
install.packages('toolStability', dependencies=TRUE)
# Install development version from Github
devtools::install_github("Illustratien/toolStability")Then the package can be loaded using the function
Welcome to toolStability
This is an r package for calculating parametric, non-parametric, and probabilistic stability indices.
Structure overview of toolStability
toolStability contains different functions to calculate stability indices, including:
- adjusted coefficient of variation
- coefficient of determination
- coefficient of regression
- deviation mean squares
- ecovalence
- environmental variance
- genotypic stability
- genotypic superiority measure
- safety first index
- stability variance
- variance of rank
Build-in data set
The default data set Data is the subset of \(APSIM\) simulated wheat data set, which
includes 5 genotypes in 4 locations for 4 years, with 2 nitrogen
application rates, 2 sowing dates, and 2 \(CO_2\) levels of treatments (Casadebaig et al.,
2016). Full dataset used in the publication see here click.
Data in this package is a data frame with 640
observations and 8 variables.
| Parameters | Number | Description |
|---|---|---|
| Trait | Wheat yield (\(kg~ha^{-1}\)). | |
| Genotype | 5 | varieties. |
| Environment | 128 | unique combination of environments for each genotype. |
| Year | 4 | years. |
| Sites | 4 | locations. |
| Nitrogen | 2 | nitrogen application levels. |
| \(CO_2\) | 2 | \(CO_2\) concentration levels. |
| Sowing | 2 | sowing dates. |
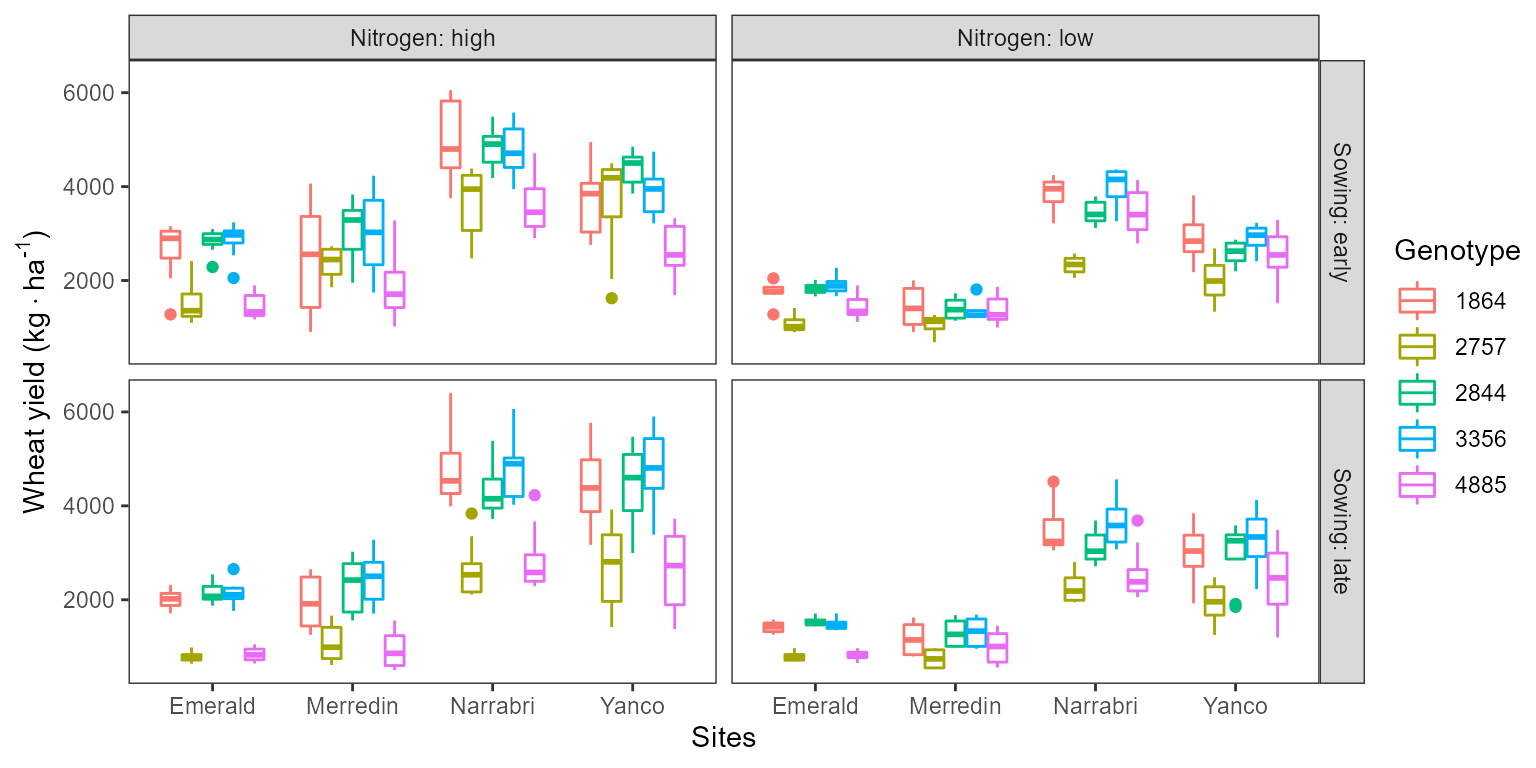
Example of environmental effects on wheat yield.
Tutorial
- Data preparation
In order to calculate stability index, you will need to prepare a
data frame with 3 columns containing trait,
genotype, and environment.
-
trait: numeric and continuous, trait value to be analyzed. -
genotype: character or factor, labeling different genotypic varieties. -
environment: character or factor, labeling different environments.
- Input formats of function
Most of the functions in the package work with the following format:
function(data = Data,
trait = "Trait_Column_Name",
genotype = "Genotype_Column_Name",
environment = "Environment_Column_Name")For calculation of probabilistic stability index
safety_first_index, an additional parameter
lambda is required.
lambda: minimal acceptable value of trait
that the user expected from crop across environment.
lambda should between the range of trait
value.
Under the assumption of trait is normally distributed,
safety first index is calculated based on the probability of trait below
lambda across the environment for each
genotype.
- Function Features
Functiontable_stability generates the summary table
containing all the stability indices in the package for every genotypes,
also including the mean trait value and normality check
results for the trait of each genoytpe across
all the enviornment.
User can specify the interested combination of environments by
entering a vector of column names which containing environmental
factors. Option normalize = TRUE allow user to compare
between different stability indices. Option
unit.correct = TRUE is designed for getting the square root
value of stability indices which have the squared unit of
trait. For function ecovalence, option
modify = TRUE takes the number of environments into account
and make modified ecovalence comparable between different
number of environments.
Examples
rm(list=ls())
library(toolStability)
### load data
data("Data")
### check the structure of sample dataset
### be sure that the trait is numeric!!!
dplyr::glimpse(Data)
#> Rows: 640
#> Columns: 8
#> $ Genotype <fct> 1864, 1864, 1864, 1864, 1864, 1864, 1864, 1864, 1864, 1864~
#> $ Yield <dbl> 1278.6, 1746.0, 1753.9, 1851.8, 2176.6, 2783.3, 3113.3, 27~
#> $ Environment <fct> 1959 Emerald low control early, 1960 Emerald low control e~
#> $ Years <int> 1959, 1960, 1961, 1962, 1959, 1960, 1961, 1962, 1959, 1960~
#> $ Sites <fct> Emerald, Emerald, Emerald, Emerald, Yanco, Yanco, Yanco, Y~
#> $ Nitrogen <fct> low, low, low, low, low, low, low, low, low, low, low, low~
#> $ CO2 <fct> control, control, control, control, control, control, cont~
#> $ Sowing <fct> early, early, early, early, early, early, early, early, ea~
### calculate ecovalence for all genotypes
single.index.ecovalence <- ecovalence(data = Data,
trait = 'Yield',
genotype = 'Genotype',
environment = 'Environment',
unit.correct = FALSE,
modify = FALSE)
### check the structure of result
dplyr::glimpse(single.index.ecovalence)
#> Rows: 5
#> Columns: 3
#> $ Genotype <fct> 1864, 2757, 2844, 3356, 4885
#> $ Mean.Yield <dbl> 2878.070, 1913.365, 2911.395, 3038.426, 2024.919
#> $ ecovalence <dbl> 17802705, 27718900, 9365241, 12698454, 24133596
### calculate modified ecovalence for all genotypes
single.index.ecovalence.modified <- ecovalence(data = Data,
trait = 'Yield',
genotype = 'Genotype',
environment = 'Environment',
unit.correct = FALSE,
modify = TRUE)
### check the structure of result
dplyr::glimpse(single.index.ecovalence.modified)
#> Rows: 5
#> Columns: 3
#> $ Genotype <fct> 1864, 2757, 2844, 3356, 4885
#> $ Mean.Yield <dbl> 2878.070, 1913.365, 2911.395, 3038.426, 2024.919
#> $ ecovalence.modified <dbl> 139083.63, 216553.91, 73165.94, 99206.67, 188543.72
### calculate all stability indices for all genotypes
summary.table <- table_stability(data = Data,
trait = 'Yield',
genotype = 'Genotype',
environment = 'Environment',
lambda = median(Data$Yield),
normalize = FALSE,
unit.correct = FALSE)
#> Warning in table_stability(data = Data, trait = "Yield", genotype = "Genotype", :
#> All of your genotypes didn't pass the Shapiro normality test!
#> Safety_first Index may not be accurate.
#### warning message means your data structure is not distributed as normal distribution
#### check the structure of result
dplyr::glimpse(summary.table)
#> Rows: 5
#> Columns: 15
#> $ Genotype <fct> 1864, 2757, 2844, 3356, 4885
#> $ Mean.Yield <dbl> 2878.070, 1913.365, 2911.395, 3038.4~
#> $ Normality <lgl> FALSE, FALSE, FALSE, FALSE, FALSE
#> $ Safety.first.index <dbl> 0.3523378, 0.6665326, 0.3242059, 0.2~
#> $ Coefficient.of.determination <dbl> 0.9398731, 0.8270000, 0.9485154, 0.9~
#> $ Coefficient.of.regression <dbl> 1.1596475, 0.8552736, 1.0316158, 1.1~
#> $ Deviation.mean.squares <dbl> 108789.28, 193280.79, 73052.65, 8677~
#> $ Environmental.variance <dbl> 1809327, 1117230, 1418923, 1630384, ~
#> $ Genotypic.stability <dbl> 29248135, 24360429, 14583562, 214768~
#> $ Genotypic.superiority.measure <dbl> 89307.69, 1004043.78, 70091.10, 3048~
#> $ Variance.of.rank <dbl> 1.770116, 2.281250, 1.561946, 1.7913~
#> $ Stability.variance <dbl> 173448.30, 303582.09, 62720.42, 1064~
#> $ Adjusted.coefficient.of.variation <dbl> 50.31578, 47.87130, 44.31829, 46.565~
#> $ Ecovalence <dbl> 17802705, 27718900, 9365241, 1269845~
#> $ Ecovalence.modified <dbl> 139083.63, 216553.91, 73165.94, 9920~
### calculate all stability indices for all genotypes
normalized.summary.table <- table_stability(data = Data,
trait = 'Yield',
genotype = 'Genotype',
environment = 'Environment',
lambda = median(Data$Yield),
normalize = TRUE,
unit.correct = FALSE)
#> Warning in table_stability(data = Data, trait = "Yield", genotype = "Genotype", :
#> All of your genotypes didn't pass the Shapiro normality test!
#> Safety_first Index may not be accurate.
#### warning message means your data structure is not distributed as normal distribution
#### check the structure of result
dplyr::glimpse(normalized.summary.table)
#> Rows: 5
#> Columns: 15
#> $ Genotype <fct> 1864, 2757, 2844, 3356, 4885
#> $ Mean.Yield <dbl> 2878.070, 1913.365, 2911.395, 3038.4~
#> $ Normality <lgl> FALSE, FALSE, FALSE, FALSE, FALSE
#> $ Safety.first.index <dbl> 0.85683453, 0.00000000, 0.93355270, ~
#> $ Coefficient.of.determination <dbl> 0.07112157, 1.00000000, 0.00000000, ~
#> $ Coefficient.of.regression <dbl> 0.0000000, 0.9787025, 0.4116811, 0.1~
#> $ Deviation.mean.squares <dbl> 0.7027599, 0.0000000, 1.0000000, 0.8~
#> $ Environmental.variance <dbl> 0.0000000, 0.9389617, 0.5296575, 0.2~
#> $ Genotypic.stability <dbl> 0.0000000, 0.3333003, 1.0000000, 0.5~
#> $ Genotypic.superiority.measure <dbl> 0.9395799, 0.0000000, 0.9593184, 1.0~
#> $ Variance.of.rank <dbl> 0.8095988, 0.3420919, 1.0000000, 0.7~
#> $ Stability.variance <dbl> 0.5402844, 0.0000000, 1.0000000, 0.8~
#> $ Adjusted.coefficient.of.variation <dbl> 0.0000000, 0.4075840, 1.0000000, 0.6~
#> $ Ecovalence <dbl> 0.5402844, 0.0000000, 1.0000000, 0.8~
#> $ Ecovalence.modified <dbl> 0.5402844, 0.0000000, 1.0000000, 0.8~
### compare the result from summary.table and normalized.summary.table
### calculate the stability indices only based only on CO2 and Nitrogen environments
summary.table2 <- table_stability(data = Data,
trait = 'Yield',
genotype = 'Genotype',
environment = c('CO2','Nitrogen'),
lambda = median(Data$Yield),
normalize = FALSE,
unit.correct = FALSE)
#> Warning in table_stability(data = Data, trait = "Yield", genotype = "Genotype", :
#> All of your genotypes didn't pass the Shapiro normality test!
#> Safety_first Index may not be accurate.
#### check the structure of result
dplyr::glimpse(summary.table2)
#> Rows: 5
#> Columns: 15
#> $ Genotype <fct> 1864, 2757, 2844, 3356, 4885
#> $ Mean.Yield <dbl> 2878.070, 1913.365, 2911.395, 3038.4~
#> $ Normality <lgl> FALSE, FALSE, FALSE, FALSE, FALSE
#> $ Safety.first.index <dbl> 0.3523378, 0.6665326, 0.3242059, 0.2~
#> $ Coefficient.of.determination <dbl> 0.161086973, 0.138169855, 0.28644744~
#> $ Coefficient.of.regression <dbl> 1.1791003, 0.8614393, 1.3780191, 1.3~
#> $ Deviation.mean.squares <dbl> 1517867.6, 962862.6, 1012476.1, 1269~
#> $ Environmental.variance <dbl> 1809327, 1117230, 1418923, 1630384, ~
#> $ Genotypic.stability <dbl> 213741097, 130745446, 161091101, 189~
#> $ Genotypic.superiority.measure <dbl> 3688981, 6251668, 3333615, 3180826, ~
#> $ Variance.of.rank <dbl> 2644.454, 1623.286, 2007.764, 2479.2~
#> $ Stability.variance <dbl> 2025117, 1102709, 1229740, 1636649, ~
#> $ Adjusted.coefficient.of.variation <dbl> 50.31578, 47.87130, 44.31829, 46.565~
#> $ Ecovalence <dbl> 192140367, 121852817, 131532582, 162~
#> $ Ecovalence.modified <dbl> 1501096.6, 951975.1, 1027598.3, 1269~
### compare the result from summary.table and summary.table2
### see how the choice of environments affect the dataEquation of stability indices
Let \(X_{ij}\) represent the observed mean value of the genoptype \(i\) in environment \(j\), and let \(\bar{X_{i.}}\), \(\bar{X_{.j}}\) and \(\bar{X_{..}}\) denote the marginal means of genotype \(i\), environment \(j\) and the overall mean, respectively.
adjusted coefficient variation
Adjusted coefficient of variation (Döring & Reckling, 2018) is calculated based on regression function. Variety with low adjusted coefficient of variation is considered as stable. Under the linear model
\[v_{i} = a + b~m_{i}\] where \(v_{i}\) is the \(log_{10}\) of phenotypic variance and \(m_{i}\) is the \(log_{10}\) of phenotypic mean. \[\widetilde{c_{i}} = \frac{1}{\widetilde{\mu_{i}}} \left [ 10^{(2-b)~ m_{i} +(b-2)~ \bar{m}+v_{i}} \right ] ^{0.5} \times 100 \% \]
coefficient of determination
Coefficient of determination (Pinthus, 1973) is calculated based on regression function. Variety with low coefficient of determination is considered as stable. Under the linear model \[Y_{ij} =\mu + \beta_{i}~ e_{j} + g_{i} + d_{ij}\] where \(Y_{ij}\) is the observed mean value of \(i^{th}\) genotype in the \(j^{th}\) environment; \(g_{i}\), \(e_{j}\) and \(\mu\) denoting genotypic mean, environmental mean and overall population mean, respectively.
The effect of GE-interaction may be expressed as: \[(ge)_{ij} = \beta_{i}~ e_{j} + d_{ij}\] where \(\beta_{i}\) is coefficient of regression and \(d_{ij}\) is deviation from regression.
For \(s^{2}_{di}\) and \(S_{xi}^{2}\), see \(deviation~mean~squares\) and \(environmental~ variance\) for details.
Coefficient of determination may be expressed as: \[r_{i}^{2} = 1 - \frac{s_{di}^{2}}{s_{xi}^{2}} \]
where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) and \(\bar{X_{.j}}\) denoting marginal means of genotype \(i\) and environment \(j\), respectively. \(\bar{X_{..}}\) denote the overall mean of \(X\).
coefficient of regression
Coefficient of regression (Finlay & Wilkinson, 1963) is calculated based on regression function. Variety with low coefficient of regression is considered as stable. Under the linear model \[Y_{ij} =\mu + \beta_{i}~ e_{j} + g_{i} + d_{ij}\] where \(Y_{ij}\) is the observed mean value of \(i^{th}\) genotype in the \(j^{th}\) environment; \(g_{i}\), \(e_{j}\) and \(\mu\) denoting genotypic mean, environmental mean and overall population mean, respectively.
The effect of GE-interaction may be expressed as: \[(ge)_{ij} = \beta_{i}~ e_{j} + d_{ij}\] where \(\beta_{i}\) is coefficient of regression and \(d_{ij}\) is deviation from regression.
Coefficient of regression may be expressed as: \[ b_{i}=1 + \frac{\sum_{j}(X_{ij} -\bar{X_{i.}}-\bar{X_{.j}}+\bar{X_{..}}) ~ (\bar{X_{.j}}- \bar{X_{..}})}{\sum_{j}(\bar{X_{.j}}-\bar{X_{..}})^{2}}\]
where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) and \(\bar{X_{.j}}\) denoting marginal means of genotype \(i\) and environment \(j\), respectively. \(\bar{X_{..}}\) denote the overall mean of \(X\). \(b_{i}\)is the estimator of \(\beta_{i}\).
deviation mean squares
Deviation mean squares (Eberhart & Russell, 1966) is calculated based on regression function. Variety with low stability variance is considered as stable.
Deviation mean squares may be expressed as: \[ s^{2}_{di} = \frac{1}{E-2} \left [ \sum_{j}~(X_{ij} - \bar{X_{i.}}- \bar{X_{.j}} + \bar{X_{..}}^{2}) - (b_{i} - 1)^{2} ~ (\bar{X_{.j}}- \bar{X_{..}})^{2} \right ] \]
where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) and \(\bar{X_{.j}}\) denoting marginal means of genotype \(i\) and environment \(j\), respectively. \(\bar{X_{..}}\) denote the overall mean of \(X\). \(b_{i}\) is the estimation of coefficient of regression.
ecovalence
Ecovalence (Wricke,
1962) is calculated based on square and sum up the
genotype–environment interaction all over the environment. Variety with
low ecovalence is considered as stable. Ecovalence is expressed as:
\[W_{i} = \sum_{j}~(X_{ij} - \bar{X_{i.}} -
\bar{X_{.j}} + \bar{X_{..}})^{2}\] To let \(W_{i}\) comparable between experiments, we
also provide the modified ecovalence (\(W_{i}'\)), whcih take the number of
environments into account. User can get (\(W_{i}'\)) by setting
modify = TRUE.
\[W_{i}' = \frac{\sum_{j}~(X_{ij} - \bar{X_{i.}} - \bar{X_{.j}} + \bar{X_{..}})^{2}}{E-1}\] where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) denoting marginal means of genotype \(i\).
environmental variance
Environmental variance (Römer, 1917) is calculated by squared and summing up all deviation from genotypic mean for each genotype. The larger the environmental variance of one genotype is, the lower the stability.
\[S_{xi}^{2} = \frac{\sum_{j}~(X_{ij} - \bar{X_{i.}})^{2}}{E-1} \] where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) denoting marginal means of genotype \(i\).
genotypic stability
Genotypic stability (Hanson, 1970) is calculated based on regression function. Variety with low stability variance is considered as stable. Under the linear model \[Y_{ij} =\mu + \beta_{i}~ e_{j} + g_{i} + d_{ij}\] where \(Y_{ij}\) is the observed mean value of \(i^{th}\) genotype in the \(j^{th}\) environment; \(g_{i}\), \(e_{j}\) and \(\mu\) denoting genotypic mean, environmental mean and overall population mean, respectively.
The effect of GE-interaction may be expressed as: \[(ge)_{ij} = \beta_{i}~ e_{j} + d_{ij}\] where \(\beta_{i}\) is coefficient of regression and \(d_{ij}\) is deviation from regression.
Genotypic stability: \[ D_{i}^{2} = \sum_{j}~(X_{ij} - \bar{X_{i.}}- b_{min}~ \bar{X_{.j}} + b_{min}~ \bar{X_{..}})^{2}\]
where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) and \(\bar{X_{.j}}\) denoting marginal means of genotype \(i\) and environment \(j\), respectively. \(\bar{X_{..}}\) denote the overall mean of \(X\).
\(b_{min}\) is the minimum value of coefficient of regression over all environments.
genotypic superiority measure
Genotypic superiority measure (Lin & Binns, 1988) is calculatd based on means square distance between maximum value of environment j and genotype i. Variety with low genotypic superiority measure is considered as stable.
\[P_{i} = \sum_{j}^{n} \frac{(X_{ij}-M_{j})^{2}}{2n}\] where \(X_{ij}\) stands for observed trait and \(M_{j}\) stands for maximum response among all genotypes in the \(j^{th}\) location.
safety first index
Safety-first index (Eskridge, 1990) is calculated based on the normality assumption of trait over the environments. Among different environments, trait below a given critical level \(\lambda\) is defined as failure of trait. Safety-first index calculating the probability of trait failure over the environment. Variety with low safety first index is considered as stable.
\[Pr~(Y_{ij} < \lambda) = \Phi \left[ (\lambda - \mu_{i})/ \sqrt \sigma_{ii} \right]\]
where \(\lambda\) is the minimal acceptable value of trait that the user expected from crop across environments. Lambda should between the range of trait value. \(\Phi\) is the cumulative distribution function of the standard normal distribution. \(\mu_{i}\) and \(\sigma_{ii}\) is the mean and variance of the system i. Under the assumption of trait is normally distributed, safety first index is calculated based on the probability of trait below lambda across the environments for each genotype.
stability variance
Stability variance (Shukla, 1972) is calculated based on linear combination of ecovalence and mean square of genotype-environment interaction. Variety with low stability variance is considered as stable.
\[ \sigma^{2}_{i} = \frac{1}{(G-1) ~ (G-2) ~ (E-1)} \left [ G ~(G-1)~ \sum_{j}~(X_{ij} - \bar{X_{i.}}-\bar{X_{.j}} + \bar{X_{..}})^{2}- \sum_{i} \sum_{j}~(X_{ij} - \bar{X_{i.}}-\bar{X_{.j}} + \bar{X_{..}})^{2} \right ] \] where \(X_{ij}\) is the observed phenotypic mean value of genotype \(i\) (i = 1,…, G) in environment \(j\) (j = 1,…,E), with \(\bar{X_{i.}}\) and \(\bar{X_{.j}}\) denoting marginal means of genotype \(i\) and environment \(j\), respectively. \(\bar{X_{..}}\) denote the overall mean of \(X\).
Negative values of stability variance is replaced with 0.
variance of rank
Variance of rank (Nassar & Hühn, 1987) is calculated based on regression function. Variety with low variance of rank is considered as stable.
Correction for each genotype \(i\) was done by subtraction of marginal genotypic mean \(\bar{X_{i.}}\) and the addition of overall mean \(\bar{X_{..}}\). \[X_{corrected~ij} = X_{ij} - \bar{X_{i.}} + \bar{X_{..}}\] Then calculated the rank all genotypes for each environment \(j\) \[r_{ij} = rank~(X_{correctedij})\]
Variance of rank is calculated as the following equation. \[S_{i}4 = \frac{\sum_{j}~(r_{ij}-\bar{r_{i.}})^{2}}{E-1}\] where \(r_{ij}\) is the rank of genotype \(i\) in environment \(j\) and \(\bar{r_{i.}}\) is the marginal rank of genotype \(i\) over environment, based on the corrected \(X_{ij}\) values.
Citing toolStability
Wang, TC., Casadebaig, P. & Chen, TW. More than 1000 genotypes are required to derive robust relationships between yield, yield stability and physiological parameters: a computational study on wheat crop. Theor Appl Genet 136, 34 (2023). https://doi.org/10.1007/s00122-023-04264-7
relavant links
* Reproducible R code for publication https://github.com/Illustratien/Wang_2023_TAAG
* Data https://doi.org/10.5281/zenodo.4729636